Learn how to plot copy number variant (CNV) on the UCSC genome browser.
Introduction
I find it useful to put a patient copy number variant (CNV) into its gene context and to compare it to a microdeletion/microduplication syndrome critical interval.
Say hypothetically, I have a patient with the following deletion:
chr7:72734455-74110000
We are wondering if this region encompass the Williams Beuren syndrome critical region (chr7:72744455-74142672)
You can find the critical region coordinates of most CNV syndromes on DECIPHER disorders.
BED format
BED format refers to a specific way of writing your data. Please read the relevant paragraph here.
In summary, the columns indicate:
- Chromosome
- Start position
- End position
- Name of CNV
- Score (put 0 here)
- Strand
- Start position
- End position
- Color (RGB)
Plot your data
Step 1. Go to UCSC and click on custom track

Step 2. Convert the coordinates of your CNV into BED format.
Our two CNVs are:
- chr7:72734455-74110000 (patient)
- chr7:72744455-74142672 (Williams Beuren syndrome)
Below are the corresponding BED format coordinates.
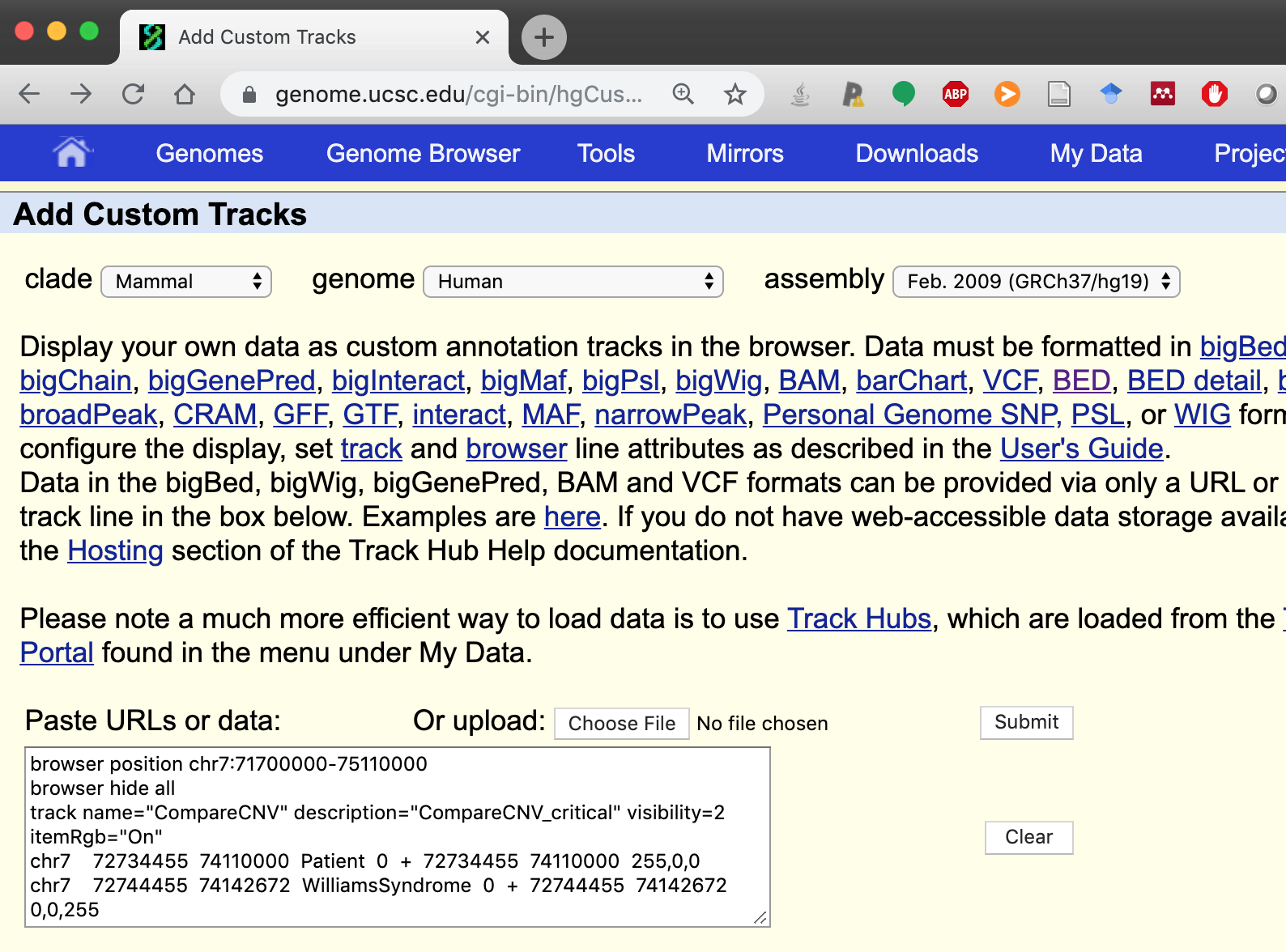
browser position chr7:71700000-75110000
browser hide all
track name="CompareCNV" description="CompareCNV_critical" visibility=2 itemRgb="On"
chr7 72734455 74110000 Patient 0 + 72734455 74110000 255,0,0
chr7 72744455 74142672 WilliamsSyndrome 0 + 72744455 74142672 0,255,0
Step 3. We copy-paste these coordinates as a custom track and click Submit.
Step 4. Click Go et voilà!!.
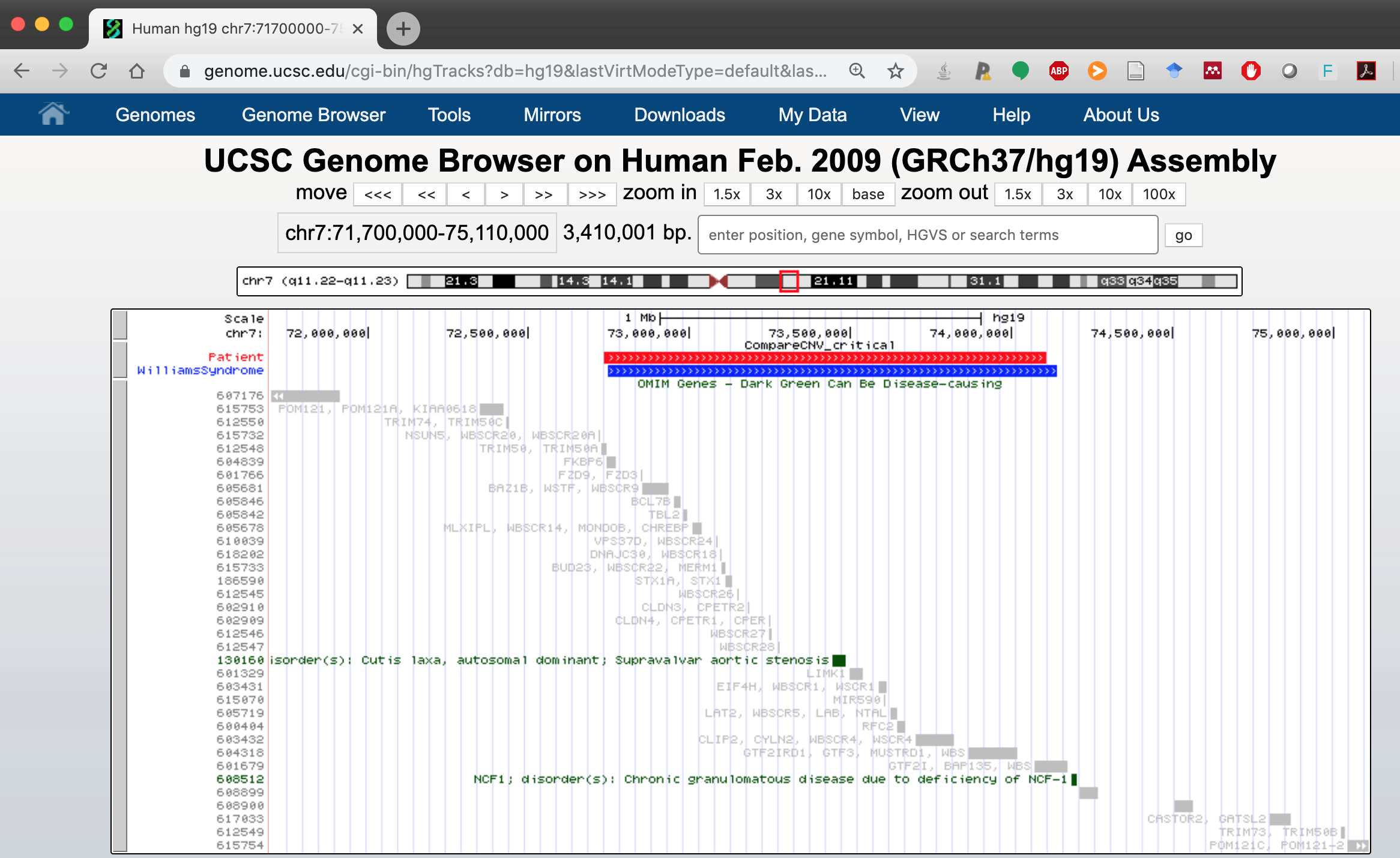
As you can see, the patient CNV is slightly shifted to the left, but it includes all the OMIM disease genes of the Williams Beuren syndrome region.
This is especially useful when a report states that a CNV “overlaps” a microdeletion/microduplication region without specifying the extent of the overlap.
It is also very useful wanting to add coordinates for recently published genomic regions and for re-analyzing old microarray results.
I hope this will help you in your practice :)
My Linh Thibodeau
comments powered by Disqus